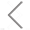Comprehensive EV Characterization
Comprehensive EV Characterization: Although FCM has been applied to analyze surface proteins of individual EVs using fluorescence threshold triggering, the minimum detectable vesicle sizes are 150-190 nm for dedicated FCM and 270-600 nm for conventional FCM, which is far from satisfaction. The NanoAnalyzer enables high-throughput multiparameter analysis of single EVs within the full size range (40~1,000 nm) and is the only commercial flow cytometric platform that can cover the entire particle size range of small EVs (40~200 nm).
Analysis of particle size and concentration
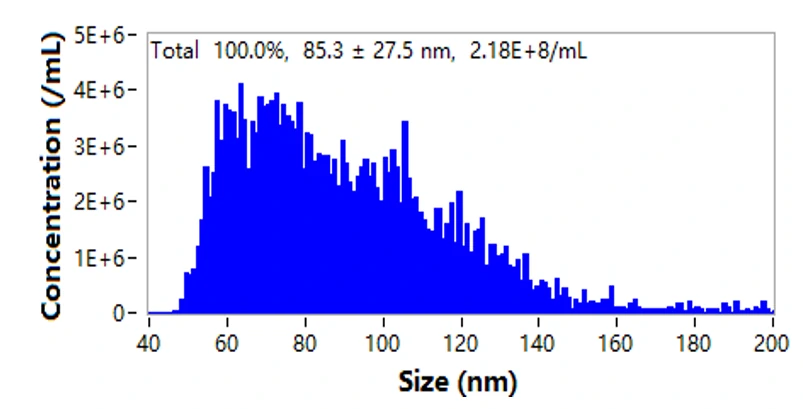
The NanoAnalyzer allows label-free analysis of individual EVs, providing particle size distribution and concentration information. Here, the particle size of EVs derived from HCT15 cells by ultracentrifugation (UC) is profiled to be 85.3 ± 27.5 nm. The particle concentration is 2.18E8 particles/mL. Meanwhile, the series dilution of the EV sample shows a very good linear correlation (R2 = 0.999).
EV purity analysis
The development of EV-based therapies and drug delivery systems requires following Good Manufacturing Practice (GMP) guidelines. Due to the complex external environment, most EV isolates contain other components (such as cell debris, protein aggregates). Therefore, the establishment of standardized EV isolation methods and the assessment of EV production is of great importance.
-
Identification of EVs by lipid-membrane dyes
-
Purity assessment of EV preparations by using Triton X-100
Detection of EV proteins
-
Profiling of EV surface proteins
-
Identification of internal proteins
Nucleic acids analysis
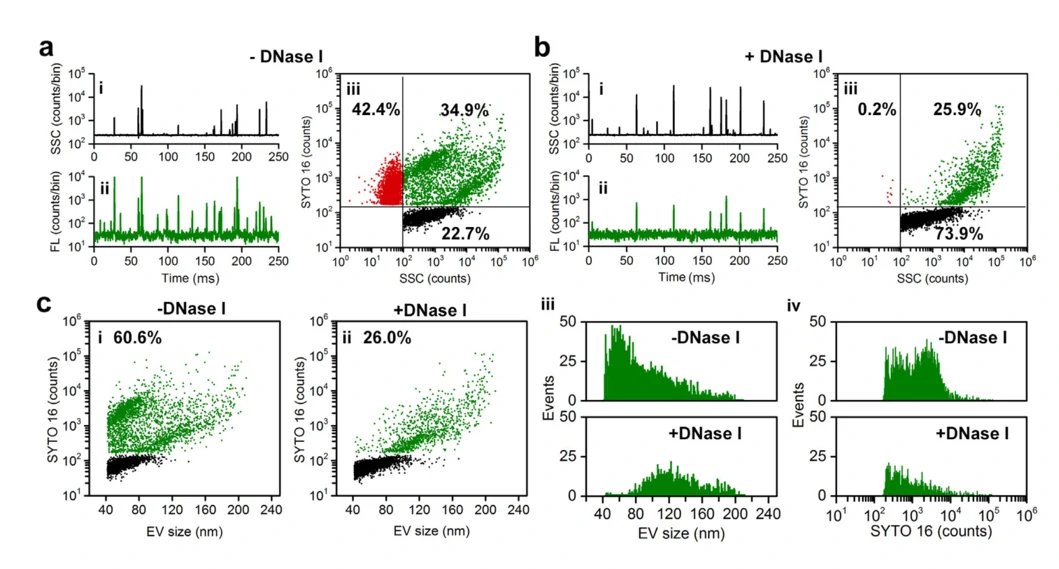
It has been demonstrated that EVs carry nucleic acids including RNA and DNA to mediate intercellular communication. Moreover, it is of great importance to understand the behaviour of these nucleic acids in EV-based nucleic acid therapeutics. Upon SYTO 16 staining and enzymatic treatment, the existence forms and distribution of DNA carried by EVs were studied at single particle level.
J Extracell Vesicles. 2022;11:e12206.
Here are some points about Comprehensive EV Characterization, read more message about NanoAnalyzer, please click here.