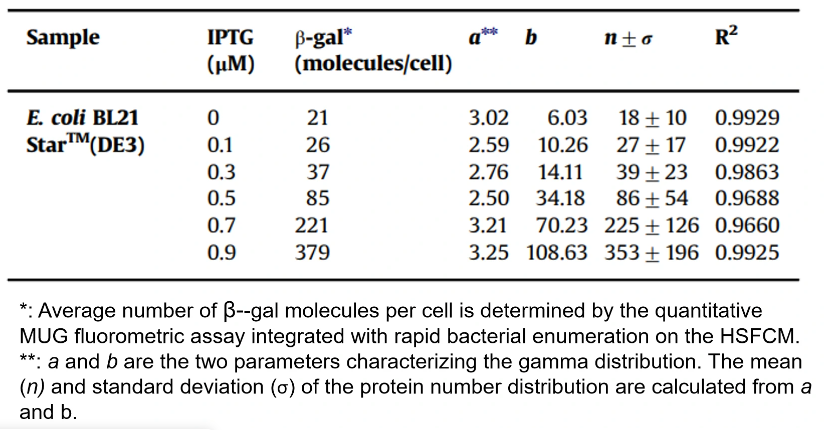
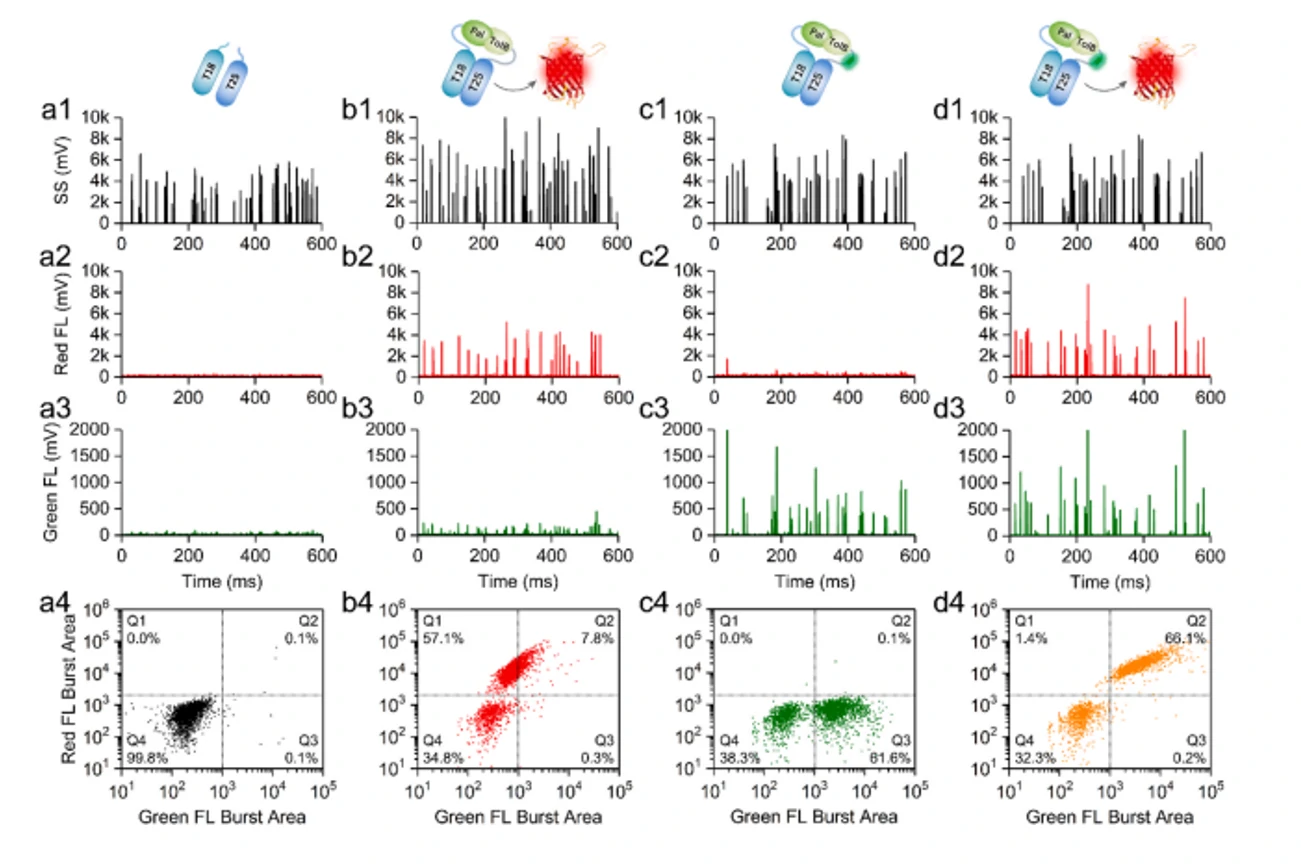
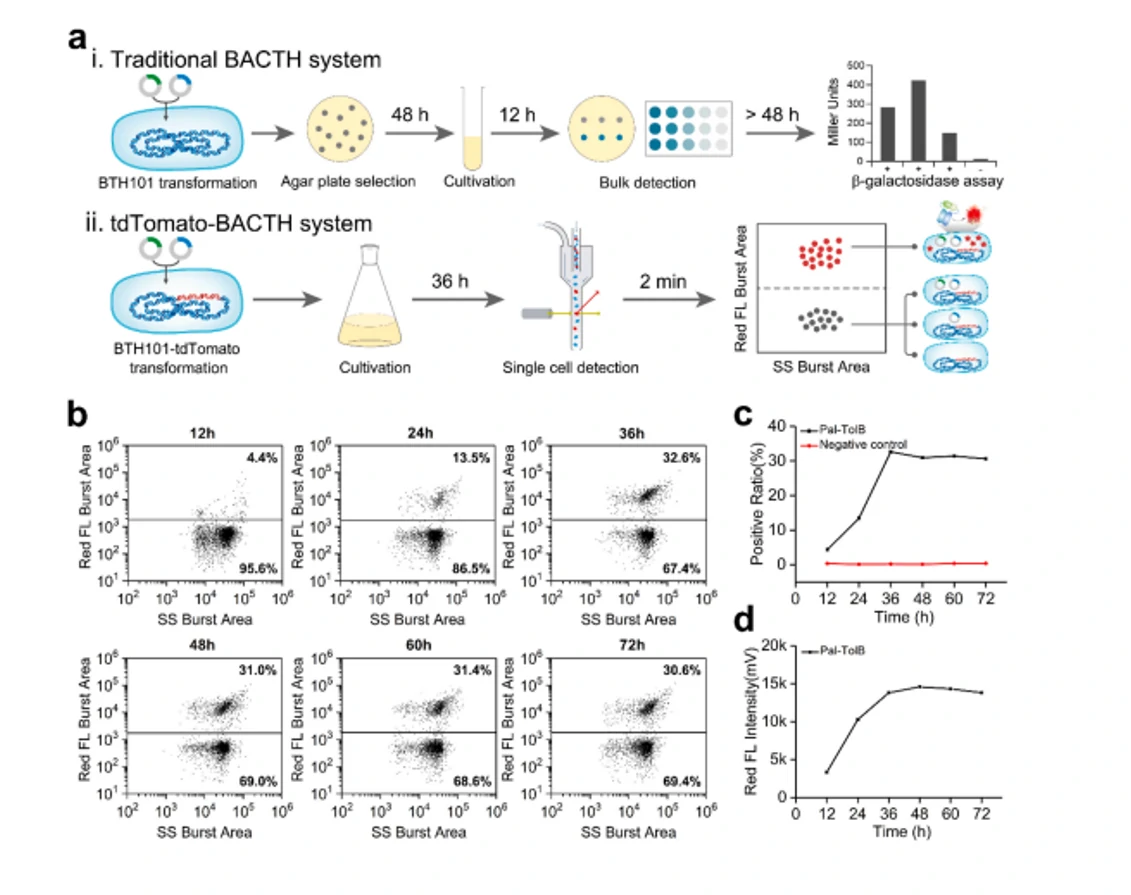
Autofluorescence Quantification of Single Bacteria
Cellular autofluorescence in the visible region can affect the sensitivity of fluorescence microscopic or flow cytometric assays by interfering with or even precluding the detection of low-level specific fluorescence. On the other hand, detection of autofluorescence can provide information for bacterial discrimination and identification. The autofluorescence detected in the green region may orginate from flavins, which comprise a category of molecules that include riboflavin (RF, vitamin B2) and its derivatives flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN). The oxidized form of flavins all share remarkably similar spectral characteristics with fluorescein isothiocyanate (FITC). Three yellow-green fluorescent Fluospheres beads with different sizes and known fluorescein equivalents were analyzed in parallel with bacterial samples to construct the calibration curve between the mean fluorescence burst area and the FITC equivalents per nanoparticle. The burst area distribution histogram of bacterial autofluorescence was then converted to the distribution of FITC equivalents per bacterial cell.
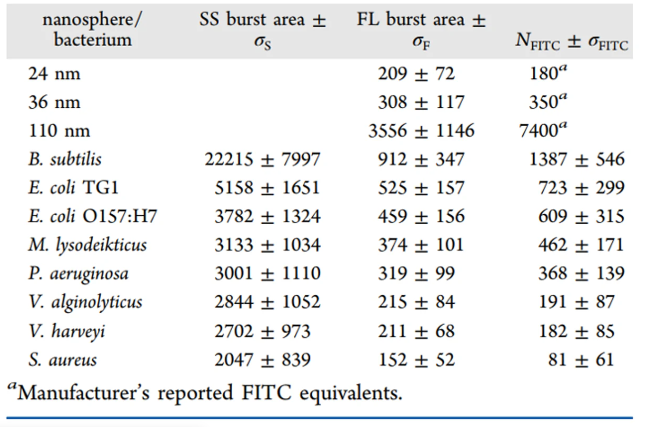
The Flow NanoAnalyzer can detect the autofluorescence of a single bacterium and demonstrate that the green autofluorescence mainly originates from the oxidized form of endogenous flavin.
Anal. Chem., 2012, 84(3), 1526-1532.