The NanoAnalyzer
Author: Mika Mika Date: May 24, 2022
The NanoAnalyzer, a nano-flow cytometry (nFCM) platform from NanoFCM, allows for the multi-parameter analysis of various biological nanoparticles, such as EVs, viruses, lipid nanoparticles, and bacteria, opening new frontiers for flow cytometry within the nanoscale. Measuring particle concentration, size distribution, and biochemical properties of single nanoparticles with high sensitivity, high resolution and at high-throughput, the NanoAnalyzer provides a powerful platform for life science research. Small EVs (<200nm) are nanoparticles that carry a very limited amount of cargo molecules such as proteins, nucleic acids and lipids. They are also highly heterogeneous in size and in contents. Therefore, a sensitive, specific, and rapid methodology is required for their characterization. To meet this demand, the NanoAnalyzer has emerged as the next-generation platform for comprehensive EV analysis.
High Sensitivity
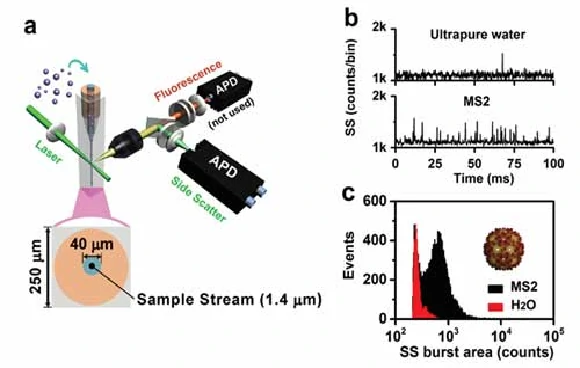
The Flow NanoAnalyzer allows you to quickly detect virus nanoparticles in a single particle without labeling. The virus used in this experiment is levivirus MS2, which is one kind of non-enveloped, spherical virion about 27 nm in diameter, and the genome is monopartite, linear ssRNA (+) about 3.5 kb in size. The signal-to-noise (S/N) ratio, which is calculated as the average burst height of all the nanoparticles detected in 1 min divided by the standard deviation of the background signal (noise), is 11 for the MS2 viruses, indicating that the Flow NanoAnalyzer provides exceptional sensitivity in discerning MS2 viruses against the background noise. This sensitivity could meet the detection demand of most virus nanoparticles in nature.
Exceptional Resolution And High Throughput
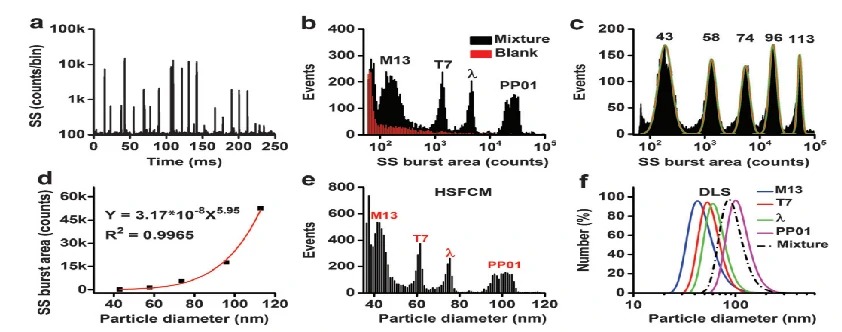
In addition to single particle characterization of viruses, the Flow NanoAnalyzer can also perform discrimination and size measurement of viruses in a mixture. A standard calibration curve is constructed with monodisperse silica nanospheres of five different diameters ranging from 43-113 nm. The SS burst areas of silica nanoparticles are plotted as a function of the diameters determined by TEM, and the SS burst area of every single virus is converted to the corresponding particle size. Employing silica nanoparticles with five different sizes to calibrate a standard curve, rapid and accurate virus size measurements with a resolution comparable to that of TEM is achieved. Particularly, with a throughput up to 10,000 particles per minute, a statistically reliable size distribution profile can be obtained in 2-3 minutes.